Research projects, objectives, collaborators
Collaborators:
- Prof. Jonathan Eggenschwiler, Department of Genetics, University of Georgia
- Prof. Karl Lechtreck, Department of Cellular Biology, University of Georgia
- Prof. Nick Galati, Department of Biology, Western Washington University
- Dr. Cecilia Lo, Department of Developmental Biology, University of Pittsburgh
Modeling and analysis of ciliary motion
We seek to develop an unbiased, computational method for analyzing ciliary motion and building a quantitative "library" of ciliary motion phenotypes. Motile respiratory cilia lining the nasal and bronchial passages beat synchronously to clear mucus and foreign matter from the respiratory tract. This mucociliary defense mechanism is essential for pulmonary health; when it breaks down, numerous ciliopathies result which can be difficult to identify without a computational detection method. Another goal is to package the analysis frameworks we develop into an open source distribution and the back-end to a web platform so other researchers can benefit from our work.
View ProjectDynamic organellar networks
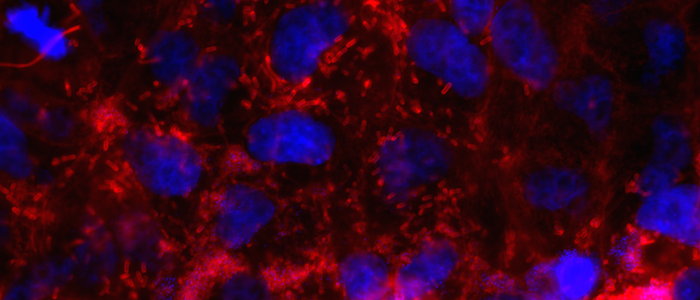
We are only beginning to scratch the surface in terms of how infections affect their host. Studying spatiotemporal changes in cellular structure as it responds to viral or bacterial infection is a crucial first step to developing therapies. We aim to develop a general and scalable graph neural network (GNN) architecture for 4D tracking of spatiotemporal evolution of tagged organelles in fluorescence microscope images, using graph theoretic techniques to embed and quantify spatiotemporal changes.
Collaborators:
- Prof. Frederick Quinn, Department of Infectious Diseases, University of Georgia
- Dr. Russell Karls, Department of Infectious Diseases, University of Georgia
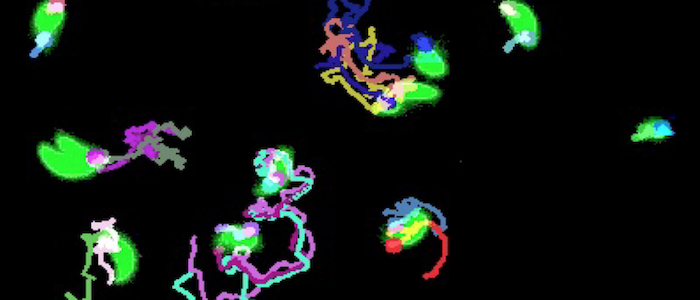
Motility and fluorescence oscillation tracking in Toxoplasma gondii
T. gondii is the causative agent of disseminated toxoplasmosis, a condition which affects over a billion people worldwide. The parasite's virulence, its ability to invade host cells, is directly linked to its motility. We examine the parasites at various stages of their lytic cycle to better understand their motility, changes in motility as a function of certain drugs, and efficacy of certain biochemical envrionments to inhibiting motility.
Collaborators:
- Prof. Silvia Moreno, Department of Cellular Biology, University of Georgia
- Prof. Gary Ward, Department of Microbiology and Molecular Genetics, University of Vermont
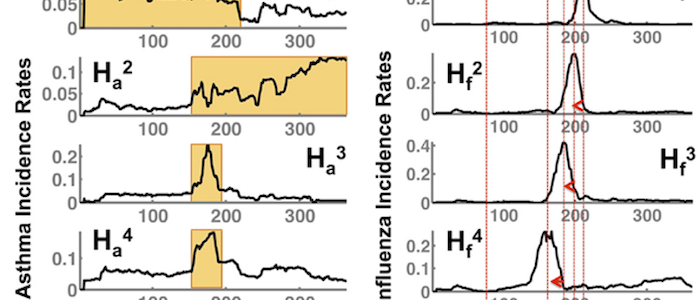
Biosurveillance and disease forecasting
The juxtaposition of big data analytics with rigorous disease modeling has driven research into scalable health analytics and predictive modeling. We explore temporal text embedding methods to detecting and predicting threats to public health that fuse "traditional" sources of health information (CDC flu reports, electronic health care reimbursement claims) with "non-traditional" sources of information (social media) to obtain nearly-real-time analytics with robust disease modeling.
Collaborators:
- Dr. Arvind Ramanathan, Data Science and Learning Division, Argonne National Laboratory
- Dr. Tim Heckman, Department of Public Health, University of Georgia
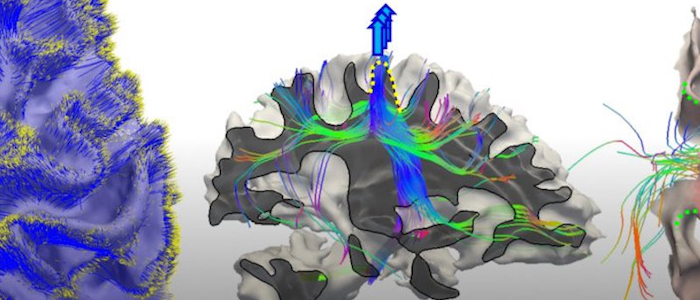
Parkinson's Disease subtyping and computational neuroscience
Parkinson's Disease (PD), and other neurodegenerative diseases, are poorly understood in terms of their progression; by the time a definitive diagnosis is made it is often too late for proactive treatments. We aim to use latent modeling and deep representation learning to characterize the spatiotemporal progression of PD and identify its subtypes.
Collaborators:
- Prof. Tianming Liu, Department of Computer Science, University of Georgia
Spatiotemporal representation learning
We are leveraging recent advances in deep neural networks and representation learning to investigate the question: how do we represent and parameterize latent representations of data with spatial and temporal components? Variational autoencoders (VAEs) use high-dimensional Gaussians to embed data, but this imposes very strong assumptions on the original format of the data, potentially warping the latent space in ways that are irrelevant to the original problem. Generative Adversarial Networks (GANs) do not impose such assumptions on the data, but analytical querying of the learned latent space is impossible. In both cases, jointly embedding spatiotemporal properties is ambiguously defined at best. Such parameterized embeddings would enable considerable advances in applied bioimaging contexts, providing tools for distinguishing phenotypes based on time lapse, appearance phenotype, and joint representation.
Open source development
Underlying all our work is a commitment to Open Science. We place heavy emphasis on communicating our results not only through clear and cogent scientific writing and speaking, but also through making our data and tools publicly available under open source licenses. We encourage everyone in the group to contribute to other open source projects. In addition to research materials, we also release course and teaching materials.