NSF CAREER objectives, recruiting, questions
CiliaWeb: Integrated platform for foundational and reproducible ciliary beat pattern analysis
Overview
The overarching goal of this project is to develop a quantitative and objective measure of ciliary motion beat patterns.
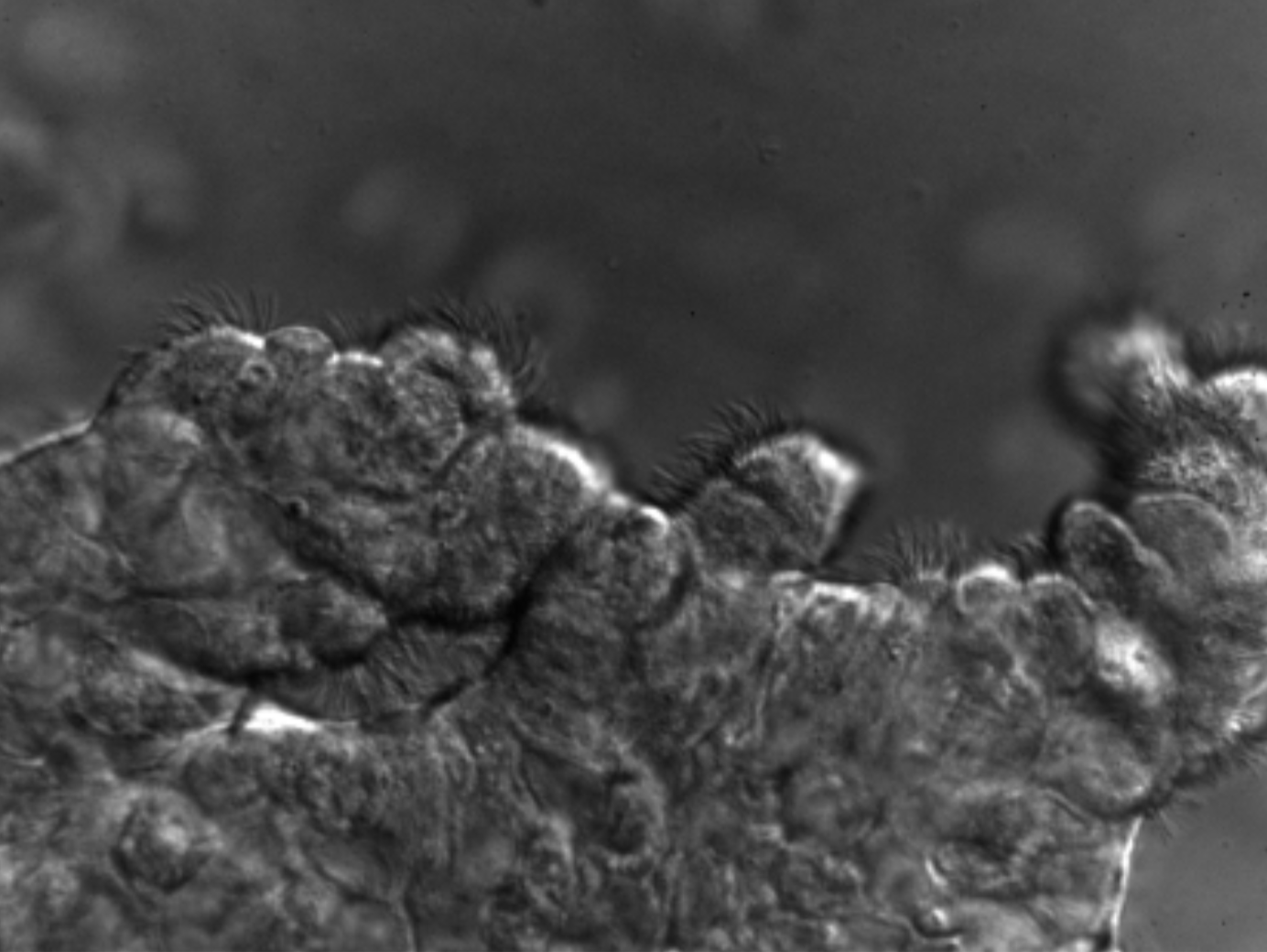
Cilia are microscopic hair-like projections from cells (Fig. 1). They beat in rhythmic patterns and perform a variety of functions, such as mucociliary clearance in the lungs and generating motility for sperm. Cilia also play sensory and signaling roles that are largely understood to involve the primary cilia, but which has also been recently implicated in motile cilia. Cilia are present along almost every major organ, including but not limited to: lungs, brain, kidneys, and throat.
Because of their presence throughout nearly all the major organs of the body, ciliopathies, or diseases of the cilia, can have wide-ranging impacts. Nearly all ciliopathies are the result of genetic mutations, but these mutations can impact either proteins that form the ciliary ultrastructure, or the intraflagellar transport (IFT) system that kicks off ciliogenesis, or the formation and maintainence system for all the cilia in the body.
Currently, the identification of ciliopathies is a process involving multiple evaluation methods. One of the more well-known methods is to compute ciliary beat frequency (CBF). CBF outside of a "normal" range (10-12Hz) tends to indicate the presence of a ciliopathy. However, there are numerous confounding variables that can present as a CBF outside that range despite the absence of a ciliopathy; conversely, certain ciliopathies can present without a detectable change in CBF. Finally, there is not a well-established methodology for rigorously evaluating CBF for a given sample. Other evaluation methods--electron microscopy to examine the ciliary ultrastructure, evaluation of ciliary beat pattern--likewise have utility but are ultimately not definitive.
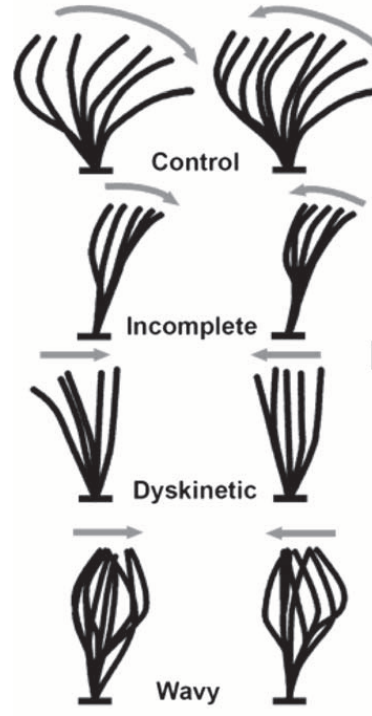
In this project, it's the ciliary beat pattern we are focused on. Researchers and clinicians generally recognize the presence of multiple discrete ciliary beat patterns (Fig. 2), but they are assessed entirely by manual inspection. Put another way, there is no rigorously-defined library of ciliary motion phenotypes.
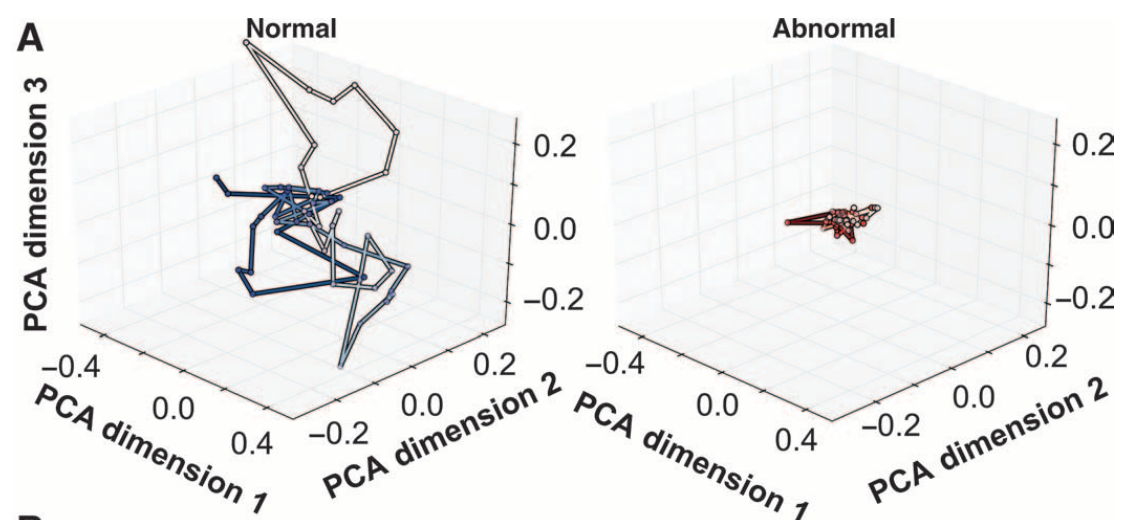
We have established some methods that have proven useful for large-scale quantitative screening and analysis of ciliary motion. We first began investigating flow-based features in our 2011 conference paper[1]. We wrapped this into a full-fledged binary classifier in our landmark 2015 Science Translational Medicine publication[2], achieving over 90% classification accuracy of normal versus abnormal ciliary motion using a support vector machine trained on two different feature sets. These feature sets revealed a low-dimensional manifold where the qualitative differences between normal and abnormal ciliary motion could be easily visualized (Fig. 3).
This paper and its findings led to a multitide of additional questions, many of which we are seeking to address as part of this project.
What constitutes a "good" subspace of ciliary motion, and how could it be used to identify ciliary motion phenotypes? As indicated in Fig. 1, it's well-established that multiple ciliary motion phenotypes exist, but we have no way of quantitatively defining them. Our 2015 paper "merely" performed normal/abnormal binary classification, ignoring any other details. There is evidence to suggest these "details" may correlate with specific genetic mutations and, therefore, provide a key for rapidly screening samples. On the advanced end, being able to "generate" samples of ciliary motion that adhere to certain motion statistics would be an incredibly useful research and diagnostic tool.
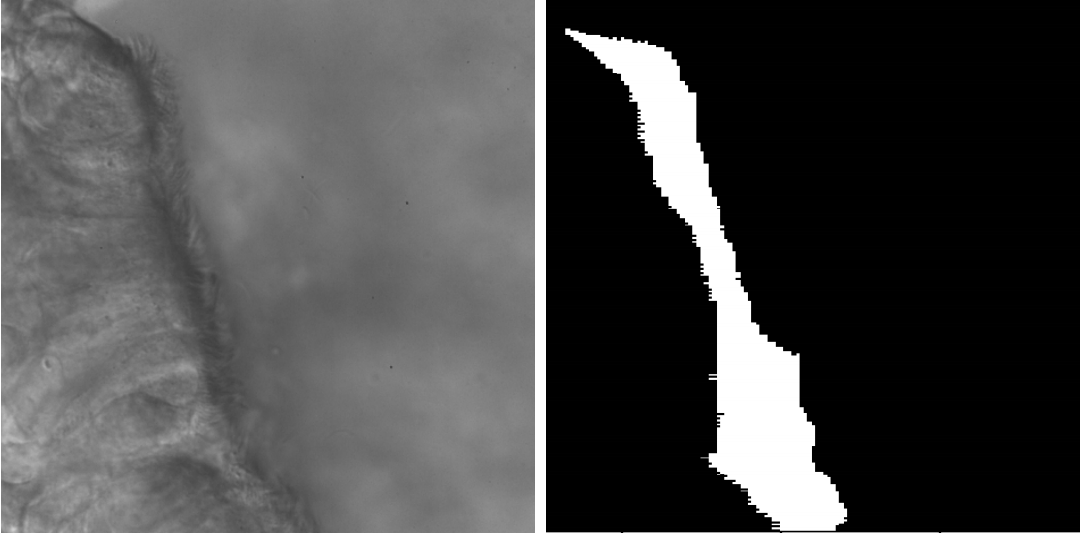
How can cilia be automatically identified in video samples? This may sound like a straightforward application of semantic video segmentation (we thought the same!), but it turns out that for cilia this is a surprisingly difficult challenge to do well[3] (Fig. 4). Identifying cilia requires clever combinations of both spatial and temporal features: cilia have a texture (sometimes), and also motion (sometimes), but neither is definitive on its own. Requiring researchers or clinicians to manually annotate samples is not only extremely labor-intensive, it's also very error-prone.
How can crowdsourcing be leveraged to include expert input while still controlling for subjectivity? One of the biggest drawbacks of the current process for ciliopathy identification is that it relies heavily on manual inspection. Nevertheless, the extensive clinical and/or laboratory training that researchers undergo should not be discounted; rather, it should be included while being kept in context. Metric learning provides a promising framework for how to approach the inclusion of manual annotations into an automated pattern recognition pipeline, but there is also physical infrastructure that is required for this: how would the interface be designed not only to encourage user recruitment and retainment, but also to incentivize high-quality feedback?
How can researchers and clinicians be encouraged to share their data and analyses? Notably, no open datasets exist around cilia research, genomic or image. Furthermore, of the little bit of software developed around ciliary motion or structure analysis, none--including ours from the 2015 paper-- is available as open source. We aim to build an integrated web platform, CiliaWeb, that is a one-stop-shop for aggregating datasets (with permission, of course), building reproducible ciliary analysis pipelines, and providing high-quality interactive visualizations and feedback. We also aim to open source the analytics and visualization engines developed as part of this project, so researchers can deploy their own versions of CiliaWeb. Finally, we're looking to generate a small amount of control and experimental genomic and image data that can be openly and publicly released for others to validate our results.
Any of this sound interesting? Read on!
Citations
Recruiting
This project will always be recruiting! If you are interested in joining some aspect of the project, regardless of your experience level, please contact me!
Frequently Asked Questions
Some questions that have both been asked, or which I'm hopefully cutting off at the pass.
I'm interested in the project, but HOLY CRAP that list of topics in the postdoc job ad is HUGE! Can I still apply?
YES! That list is meant to convey the rich variety of topics spanned by this 5-year research project, and to encourage people with a variety of backgrounds to apply! Do you have highly theoretical machine learning expertise in image representations? Apply! Are you an expert on web frameworks and user interface design? Apply! Did you do some bench work and computational analysis outside of imaging? Apply!
I want to apply but my imposter syndrome is in full swing and you'll probably get more qualified candidates than me. Why should I apply?
Speaking as someone who at times has been wholesale paralyzed by imposter syndrome, I can say with confidence: you're more qualified than you think. Like any person suffering from imposter syndrome, it's far more likely that you'll be among the most qualified. So please apply! Perhaps we could jointly plot ways to keep fooling everyone around us while we do some really neat work studying cilia!
Great! Here's my CV. When do I start getting paid?
This is a question I get a lot, and unfortunately the answer usually isn't what people are hoping. The bottom line is that I can't guarantee funding until we've at least had a chance to speak in real-time. History (not just mine) has demonstrated that while background and experience are certainly important, far more important is how well you and I (and the rest of my research group) can work together. That can only be assessed by setting up some time to chat. Ideally this would entail taking a course with me, though I know that's not always possible. Suffice to say, we should set aside time to discuss things if you absolutely have to be guaranteed funding before you'll even consider applying.
[NOTE: The above largely applies only to doctoral student applicants. Postdoctoral candidates go through a much more formal application and interview process.]
I'm not really a team player; rather, I'm a wicked good coder who will spend nights and weekends knocking down these research questions. When can I start?
Never. The tech sector and academia at large may still be enamored with the "difficult but talented" individual, but that goes against our group ethos. Plus, "talent" and "decency" are most assuredly not mutually exclusive! We absolutely work hard, but we also explicitly focus on communication, respect, and inclusiveness.
Is this project only for doctoral students and postdocs?
Not at all! I've had quite a few very successful undergraduate students conduct research with me for course credit, for CURO, and even for transitioning into Double Dawgs. Not only that, but I'm also an active participant in the UGA Young Dawgs program and recruited students from high schools in the Athens area. My only preference for recruiting undergraduates is that you start working with me in your second or third year, so we have time to develop some momentum and develop a good professional rapport.
If you're an undergraduate and want to work on this project, just shoot me an email!
I'm a life sciences student/graduate with little to no computational experience. Should I still apply?
As long as you are open to and enthusiastic about picking up at least some computational skills, then absolutely yes! You will be expected to pick up some fundamental skills in machine learning and coding, and this will certainly comprise the majority of your work, but there will also be opportunities to do some bench and wet lab work.
I have a question that isn't answered here. How do I get it answered?
Award Details
| Abstract | #1845915 |
|---|---|
| NSF Directorate | Biological Sciences (BIO) |
| NSF Division | Biological Infrastructure (DBI) |